
Breaking News
 We MUST keep talking about this, demand Voter ID. Joe Rogan and Elon Musk on Fraud
We MUST keep talking about this, demand Voter ID. Joe Rogan and Elon Musk on Fraud
 Nick Shirley exposes there are 1,200 medical transport companies in Minnesota.
Nick Shirley exposes there are 1,200 medical transport companies in Minnesota.
 Trump Explains How The US Really 'Is' NATO
Trump Explains How The US Really 'Is' NATO
 Trump Says "Investigation Of California Has Begun" As Surgical Truth-Bombs About Corruptio
Trump Says "Investigation Of California Has Begun" As Surgical Truth-Bombs About Corruptio
Top Tech News
 The First Production All-Solid-State Battery Is Here, And It Promises 5-Minute Charging
The First Production All-Solid-State Battery Is Here, And It Promises 5-Minute Charging
 See inside the tech-topia cities billionaires are betting big on developing...
See inside the tech-topia cities billionaires are betting big on developing...
 Storage doesn't get much cheaper than this
Storage doesn't get much cheaper than this
 Laser weapons go mobile on US Army small vehicles
Laser weapons go mobile on US Army small vehicles
 EngineAI T800: Born to Disrupt! #EngineAI #robotics #newtechnology #newproduct
EngineAI T800: Born to Disrupt! #EngineAI #robotics #newtechnology #newproduct
 This Silicon Anode Breakthrough Could Mark A Turning Point For EV Batteries [Update]
This Silicon Anode Breakthrough Could Mark A Turning Point For EV Batteries [Update]
 Travel gadget promises to dry and iron your clothes – totally hands-free
Travel gadget promises to dry and iron your clothes – totally hands-free
 Perfect Aircrete, Kitchen Ingredients.
Perfect Aircrete, Kitchen Ingredients.
 Futuristic pixel-raising display lets you feel what's onscreen
Futuristic pixel-raising display lets you feel what's onscreen
 Cutting-Edge Facility Generates Pure Water and Hydrogen Fuel from Seawater for Mere Pennies
Cutting-Edge Facility Generates Pure Water and Hydrogen Fuel from Seawater for Mere Pennies
Harvard scientists uncover an exploitable Achilles' heel common to most bacteria
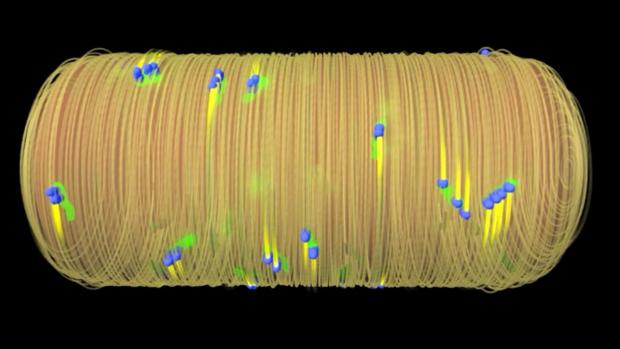
Finding ways to disarm these defenses is a key component of antibiotics, and now researchers at Harvard Medical School have identified a structural weakness that seems to be built into a range of bacterial species, potentially paving the way for a new class of widely-effective antibacterial drugs.
The new study builds on previous research into a protein named RodA. While the protein itself has long been known, in 2016 the Harvard team was the first to discover that it builds the protective cell walls of bacteria out of sugar molecules and amino acids. Since RodA belongs to the SEDS family of proteins, which is common to almost all bacteria, the team realized it was the perfect target for a far-reaching antibiotic. And on closer examination of RodA, the researchers spotted a vulnerable looking cavity on the outer surface of the protein.
"What makes us excited is that this protein has a fairly discrete pocket that looks like it could be easily and effectively targeted with a drug that binds to it and interferes with the protein's ability to do its job," says David Rudner, co-senior author of the study.
To test whether this cavity was the Achilles' heel they were looking for, the scientists altered the structure of the protein in two species of bacteria, E. coli and Bacillus subtilis. These two were chosen because they're well understood and represent the two broad classes of disease-causing bacteria, gram-positive and gram-negative.



